Breaking Down of Model Predictions for lm models
Breaking Down of Model Predictions for lm models
# S3 method for lm broken(model, new_observation, ..., baseline = 0, predict.function = stats::predict.lm)
Arguments
| model | a lm model |
|---|---|
| new_observation | a new observation with columns that corresponds to variables used in the model |
| ... | other parameters |
| baseline | the orgin/baseline for the breakDown plots, where the rectangles start. It may be a number or a character "Intercept". In the latter case the orgin will be set to model intercept. |
| predict.function | function that will calculate predictions out of model (typically |
Value
an object of the broken class
Examples
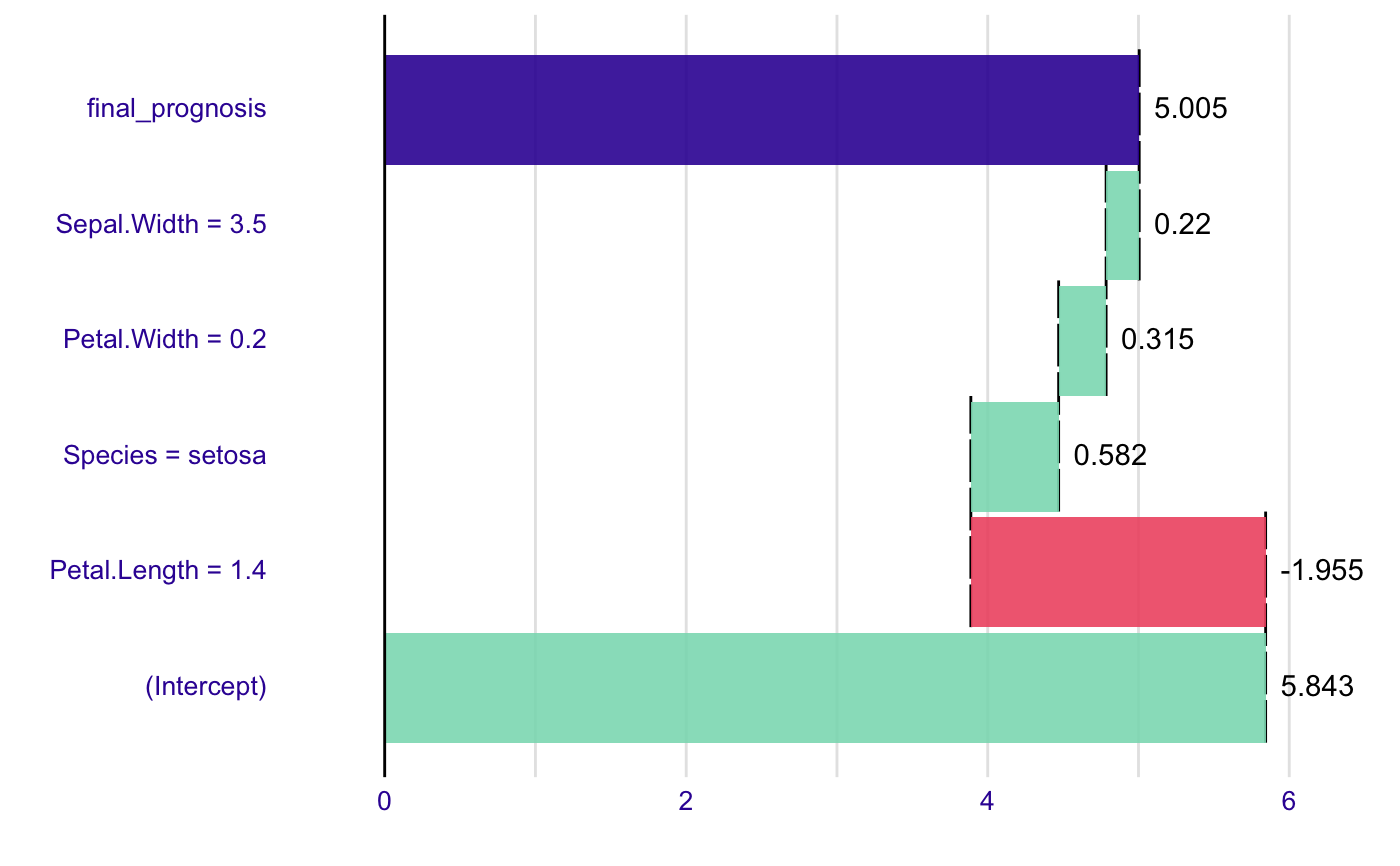
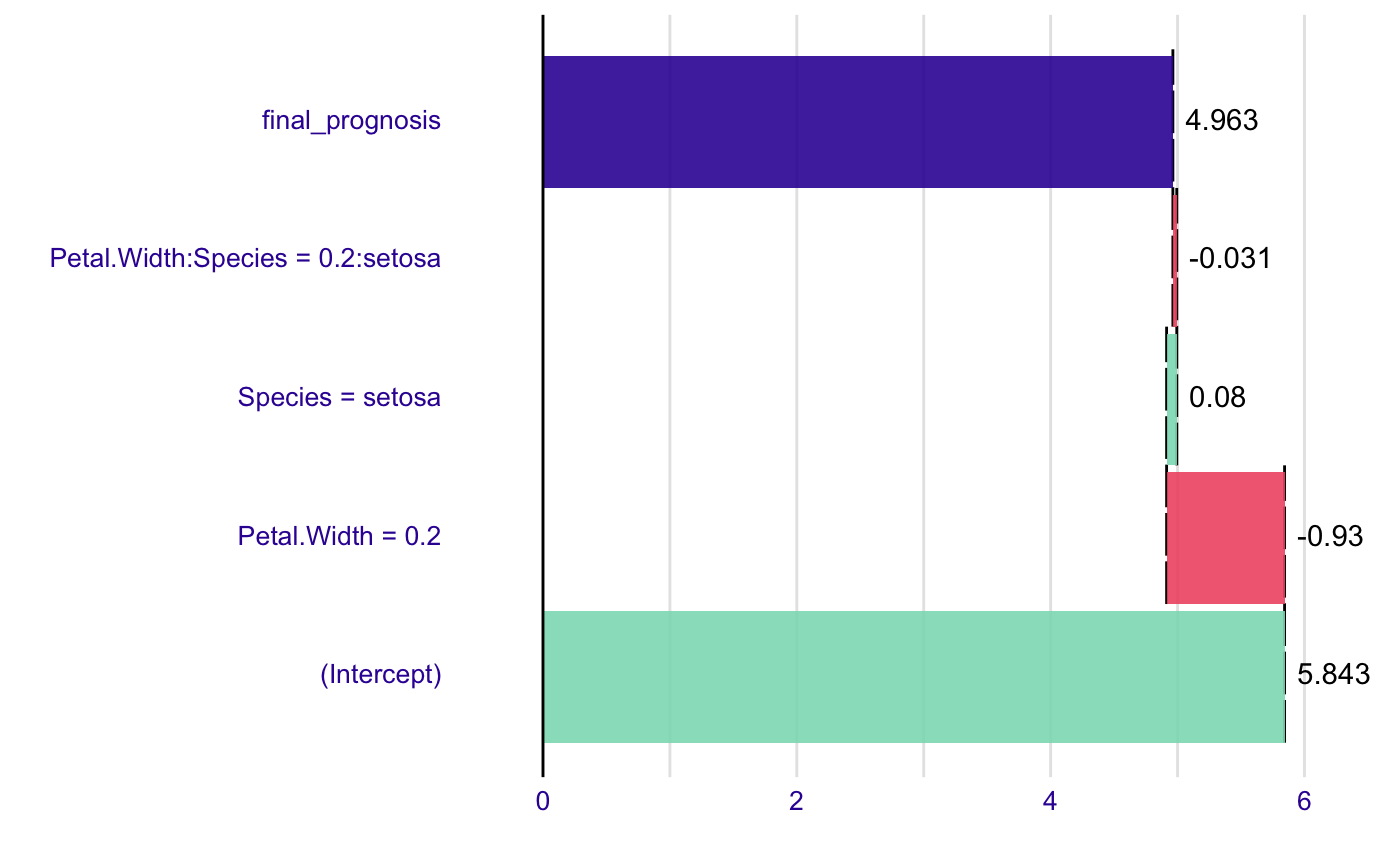
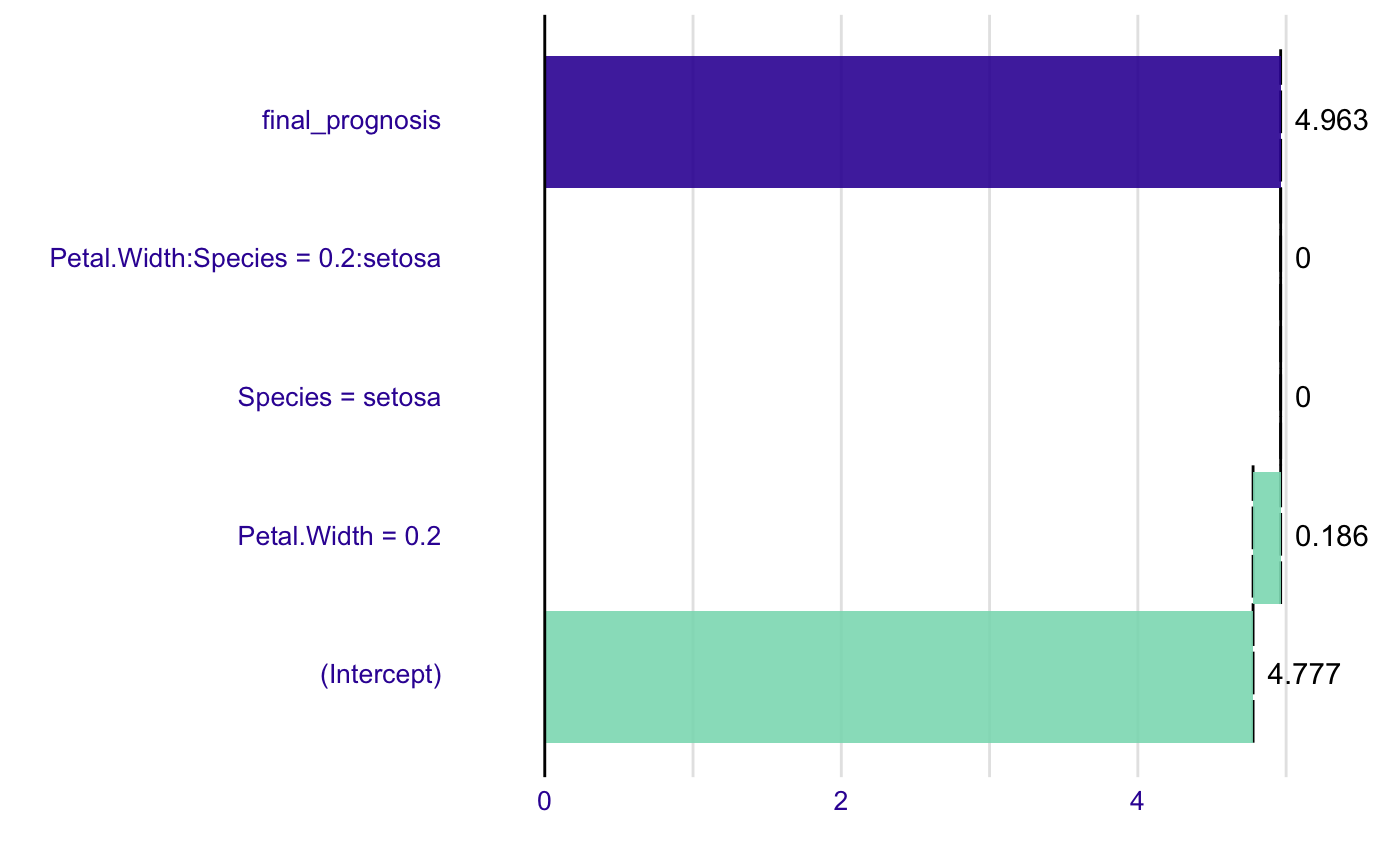
model <- lm(Sepal.Length~., data=iris) new_observation <- iris[1,] br <- broken(model, new_observation) plot(br)# works for interactions as well model <- lm(Sepal.Length ~ Petal.Width*Species, data = iris) summary(model)#> #> Call: #> lm(formula = Sepal.Length ~ Petal.Width * Species, data = iris) #> #> Residuals: #> Min 1Q Median 3Q Max #> -1.47583 -0.29891 -0.04508 0.25075 1.32892 #> #> Coefficients: #> Estimate Std. Error t value Pr(>|t|) #> (Intercept) 4.7772 0.1735 27.541 <2e-16 *** #> Petal.Width 0.9302 0.6491 1.433 0.154 #> Speciesversicolor -0.7325 0.4951 -1.480 0.141 #> Speciesvirginica 0.4922 0.5379 0.915 0.362 #> Petal.Width:Speciesversicolor 0.4962 0.7356 0.675 0.501 #> Petal.Width:Speciesvirginica -0.2793 0.6953 -0.402 0.688 #> --- #> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1 #> #> Residual standard error: 0.4789 on 144 degrees of freedom #> Multiple R-squared: 0.6768, Adjusted R-squared: 0.6656 #> F-statistic: 60.31 on 5 and 144 DF, p-value: < 2.2e-16 #>#> contribution #> (Intercept) 5.843 #> Petal.Width = 0.2 -0.930 #> Species = setosa 0.080 #> Petal.Width:Species = 0.2:setosa -0.031 #> final_prognosis 4.963 #> baseline: 0plot(br)#> contribution #> (Intercept) 4.777 #> Petal.Width = 0.2 0.186 #> Species = setosa 0.000 #> Petal.Width:Species = 0.2:setosa 0.000 #> final_prognosis 4.963 #> baseline: 0plot(br2)